Introduction
👋🏻 What is this workshop about?
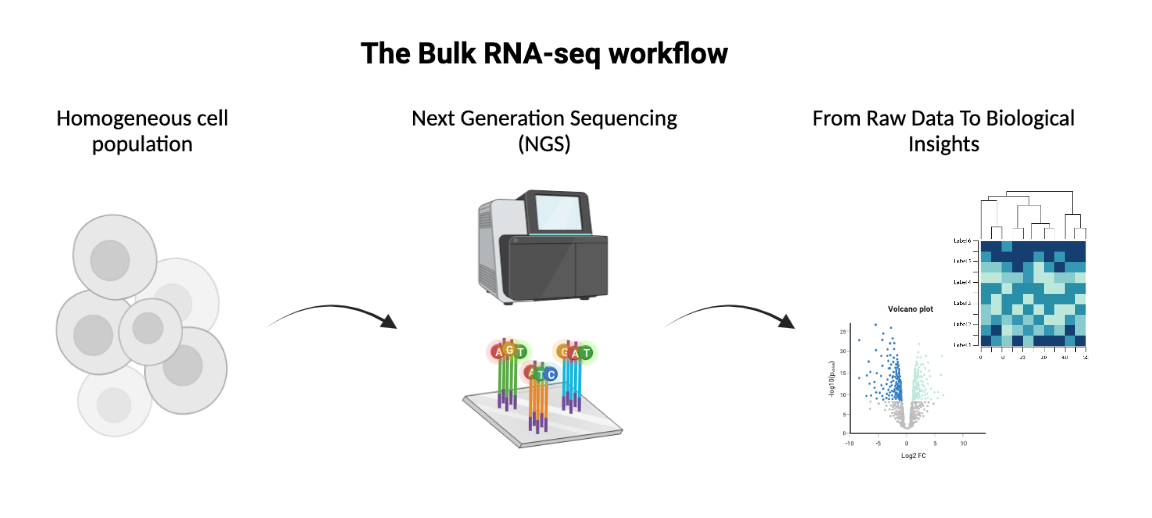
The course will give an overview on how to conceptually approach bioinformatics. During the three days of the course we will dive into a bulk RNA-seq experiment and carry out the main steps of a standard data analysis pipeline.
Program Outline
Day 1 🤓
- Setup RStudio or Posit
- Get familiar with the Posit interface
- Learn about bulk RNA-seq data processing
- Download the data needed for the workshop
Day 2 🧑🏼💻
- Learn about data normalization
- Learn about the
edgeRpackage - Explore different normalization methods
- Normalize the data with functions provided by the
edgeRpackage - Perform diagnostic and exploratory analysis on the data
Day 3 🧞♂️
- Learn about the theory behind differential expression analysis
- Perform differential expression analysis using
edgeR - Visualize the results
- Perform further downstream analysis on interesting gene groups
How To Reach Us
Jacopo Arrigoni(jacopo.arrigoni@ifom.eu)Mattia Toninelli(mattia.toninelli@ifom.eu)
Feel free to drop us an e-mail if you have any curiosity or question!
Credits
Some of the images and inspiration came from other tutorials available online, in particular this work for images and this work for some of the information regarding data normalization. All of the code present within this site was developed and is readily implementable in RStudio using RMarkdown.
License
All of the material in this course is under a Creative Commons Attribution license (CC BY 4.0) which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.